Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Virtual Trackball Rotation on PyMol
This section provides a quick introduction of Virtual Trackball Rotation model used by PyMol to rotate the molecule structure in 3-dimensions.
Once the molecule structure is displayed in the viewer window, you can rotate the molecule structure in 3-dimensions using the virtual trackball rotation model as described below.
1. The molecule structure is attached to a virtual trackball with the center of thr structure located at the center of the trackball.
2. The center of the trackball is fixed at the center of a 3-dimensional coordinate system on the screen with x-axis displayed horizontally, y-axis displayed vertically, and z-axis displayed perpendicular to the screen.
3. The trackball can be rotated by the left mouse button in 4 ways:
- Click the trackball at the center and drag is vertically to rotate it about the x-axis.
- Click the trackball at the center and drag is horizontally to rotate it about the y-axis.
- Click the trackball on the left or right edge and drag is vertically to rotate it about the z-axis.
- Click the trackball on the top or bottom edge and drag is horizontally to rotate it about the z-axis.
The following diagram (source: "Introduction to PyMOL" by DeLano Scientific LLC) shows you the virtual trackball rotation model used in PyMol:
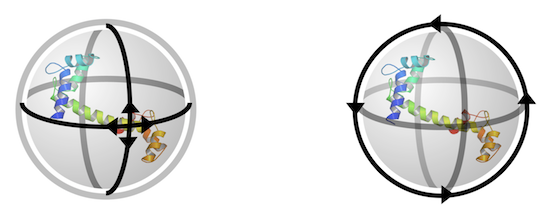
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Load Molecule from File into PyMol
►Virtual Trackball Rotation on PyMol
"load" and "delete" Commands on PyMol
"log_open" and "log_close" Commands on PyMol
Model Space and Camera Space on PyMol
"get_view" and "set_view" on PyMol
View Parameters Auto Adjusted on PyMol
Rotation with Transformation Matrix
Difference of "turn" and "rotate" Commands
Difference of "move" and "translate" Commands
"center", "zoom" and "reset" Commands
Model-to-Camera Space Coordinates Mapping
Camera-to-Model Space Coordinates Mapping
"show lines" Presentation Command
"show sticks" Presentation Command
"show spheres" Presentation Command
"show surface" Presentation Command
"show mesh" Presentation Command
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)