Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Model-to-Camera Space Coordinates Mapping
This section provides a tutorial on how to map model space coordinates to camera space coodinates manually using rotation center and rotation matrix in PyMol.
In previous tutorials, we have learned how to use "turn" and "move", to change camera viewing angle and position, which impacts how the molecule structure is mapped from the model space to the camera space.
Below is the mapping formula used by PyMol.
Assuming:
we have the following mapping relation:
We can also write the mapping relation in the vector notation as:
or:
where M is the rotation matrix:
Now let's verify this mapping formula by looking the methane molecule.
1. Load the methane molecule structure to PyMol again. You see the structure displayed at the center of the screen, which is perpendicular to the z-axis of the camera space.
PyMOL>delete all PyMOL>load Molecule-Methane.sdf CmdLoad: loaded as "Molecule-Methane".
2. Get the view mapping parameters, which include the rotation matrix and the rotation center.
PyMOL>get_view ### cut below here and paste into script ### set_view (\ 1.000000000, 0.000000000, 0.000000000,\ # M: rotation matrix 0.000000000, 1.000000000, 0.000000000,\ # M: rotation matrix 0.000000000, 0.000000000, 1.000000000,\ # M: rotation matrix 0.000000000, 0.000000000, -14.178204536,\ # D: camera rotation center 0.840320110, -1.029980063, -0.000119984,\ # B: model rotation center 11.178204536, 17.178203583, -20.000000000 ) ### cut above here and paste into script ###
3. Get coordinates of the carbon atom in the model space.
PyMOL>select carbon, id 5 Selector: selection "carbon" defined with 1 atoms. PyMOL>get_extent carbon cmd.extent: min: [ 0.840, -1.030, 0.000] cmd.extent: max: [ 0.840, -1.030, 0.000]
4. Run the formula on the carbon atom location. We get (0.000, 0.000, -14.178) in camera space. So the carbon atom should be displayed at the center of the screen.
5. Repeat the calculation on hydrogen atom #4.
PyMOL>select h4, id 4 Selector: selection "h4" defined with 1 atoms. PyMOL>get_extent h4 cmd.extent: min: [ 0.732, -1.977, 0.529] cmd.extent: max: [ 0.732, -1.977, 0.529]
We get (-0.108, -0.947, -13.649) in camera space.
You can continue on other hydrogen atoms.
The picture below shows coordinates of the carbon atom and hydrogen atom #4:
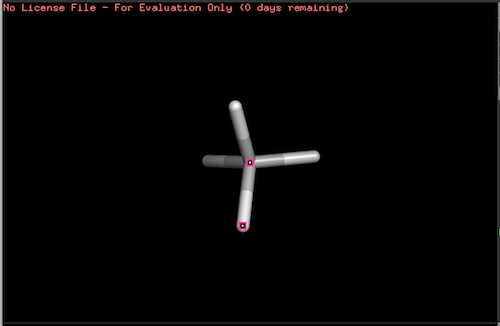
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Load Molecule from File into PyMol
Virtual Trackball Rotation on PyMol
"load" and "delete" Commands on PyMol
"log_open" and "log_close" Commands on PyMol
Model Space and Camera Space on PyMol
"get_view" and "set_view" on PyMol
View Parameters Auto Adjusted on PyMol
Rotation with Transformation Matrix
Difference of "turn" and "rotate" Commands
Difference of "move" and "translate" Commands
"center", "zoom" and "reset" Commands
►Model-to-Camera Space Coordinates Mapping
Camera-to-Model Space Coordinates Mapping
"show lines" Presentation Command
"show sticks" Presentation Command
"show spheres" Presentation Command
"show surface" Presentation Command
"show mesh" Presentation Command
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)