Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Load Molecule from File into PyMol
This section provides a tutorial on how to load a molecule structure from a file into PyMol.
In order to visualize a molecule in PyMol, you need to load the molecule structure from a file first.
1. Start PyMol and click the "File > Open" menu. You see the file open dialog box.
2. Find and select the "Molecule-HY-001.sdf" file, which stores a molecule structure in a SDF (Structure Data File) file.
3. Click "Open" to load the molecule structure from the file into PyMol. You see the molecule structure is displayed in the viewer area.
Now you can use PyMol commands or mouse clicks to control how the molecule structure is displayed.
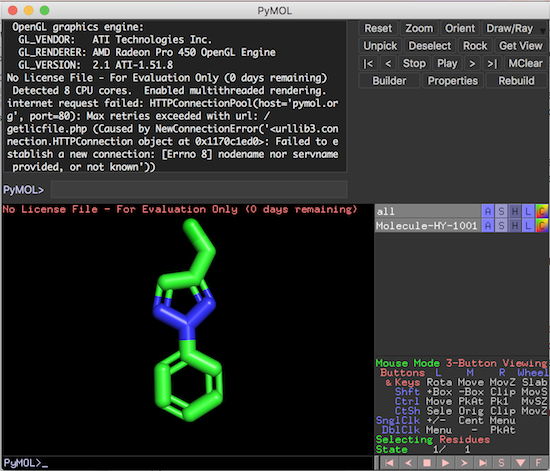
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
►Load Molecule from File into PyMol
Virtual Trackball Rotation on PyMol
"load" and "delete" Commands on PyMol
"log_open" and "log_close" Commands on PyMol
Model Space and Camera Space on PyMol
"get_view" and "set_view" on PyMol
View Parameters Auto Adjusted on PyMol
Rotation with Transformation Matrix
Difference of "turn" and "rotate" Commands
Difference of "move" and "translate" Commands
"center", "zoom" and "reset" Commands
Model-to-Camera Space Coordinates Mapping
Camera-to-Model Space Coordinates Mapping
"show lines" Presentation Command
"show sticks" Presentation Command
"show spheres" Presentation Command
"show surface" Presentation Command
"show mesh" Presentation Command
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)