Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
"get_area" - Surface Areas of Atoms
This section provides a tutorial on how to use the 'get_area' command to measure surface areas of atoms.
PyMol allows you to measure surface areas of the atoms, using the "get_area" command.
For example, the following commands print areas of the carbon atom a hydrogen atom of the methane molecule:
PyMOL># load the carbon atom PyMOL>load Carbon-Atom.sdf PyMOL># add 4 hydrogen atoms to create methane PyMOL>h_add PyMOL># print surface area of the carbon atom PyMOL>get_area id 1 cmd.get_area: 15.171 Angstroms^2. PyMOL># print surface area of a hydrogen atom PyMOL>get_area id 2 cmd.get_area: 8.258 Angstroms^2.
Based the above output, we can calculate the radius of each atom, using the sphere surface area formula: area = 4 * PI * radius * radius.
PyMOL>import math PyMOL>print str( math.sqrt(15.171/(4*3.14159)) ) 1.098758772689268 PyMOL>print str( math.sqrt(8.258/(4*3.14159)) ) 0.8106487001768432
Now we can calculate the distance between these 2 atoms. Then we can compare the distance with their radiuses and see how much they overlap with each other:
PyMOL>get_distance id 1, id 2 cmd.get_distance: 1.090 Angstroms.
If our calculation is correct, half of the hydrogen atom is overlapped with the carbon atom. This can be verified by displaying atoms as spheres.
PyMOL>show spheres, id 1 PyMOL>show spheres, id 2
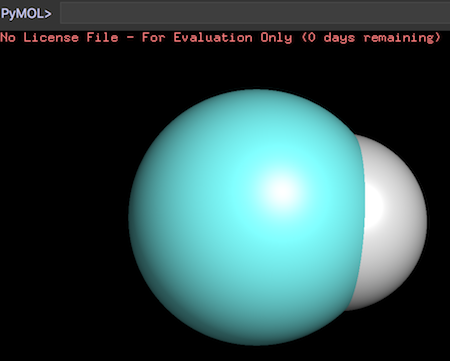
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
"get_extent" - Picked Atom Location
"label" - Generate Labels on Atoms
Distance between Atoms in PyMol
Angle Formed by 3 Atoms in PyMol
Dihedral Angle Formed by 3 Atoms in PyMol
Use Selection Expressions in PyMol
►"get_area" - Surface Areas of Atoms
Surface Area of Entire Molecule
"get_position" - Viewing Center
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)