Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Substructure Selection Visualization in PyMol
This section provides a tutorial on how to visualize a substructure selected from a molecule differently than the entire molecule structure in PyMol.
Once a selection is created, you visualize it differently than the whole molecule by specifying the selection name in the "show" command:
show representation_name, selection_name
Let's create the "ring" selection again by running all commands recorded in the log file.
PyMOL>@Selection-Create-Ring.pml ... PyMOL>hide all PyMOL># show the molecule in "lines" representation PyMOL>show lines PyMOL># show the selection in "sticks" representation PyMOL>show sticks, ring
Now the molecule structure is visualized with 2 representations superposed over each other.
- "lines" representation of the entire molecule.
- "sticks" representation of the "ring" selection.
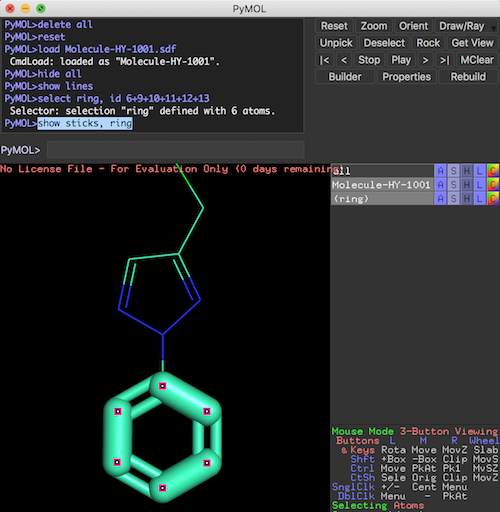
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Create Selection with Mouse in PyMol
►Substructure Selection Visualization in PyMol
Modify Molecule Structure in PyMol
Export Molecule Substructure in PyMol
Create Methane Molecule in PyMol
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)