Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
"remove pk*" and "remove_picked" Commands
This section provides a tutorial on how to remove atoms from a molecule in PyMol using picked atoms as special selections, pk1, pk2, pk3 and pk4.
Once atoms are picked and labeled as "pk1", "pk2", "pk3" and "pk4" selections, you can use "remove pk*" or "remove_picked" commands to remove picked atoms from the molecule structure. For example:
PyMOL># remove the first picked atom PyMOL>remove pk1 PyMOL># remove the all picked atoms PyMOL>remove_picked
You can also click "Build > Remove (pk1)" from the menu to remove the first picked atom.
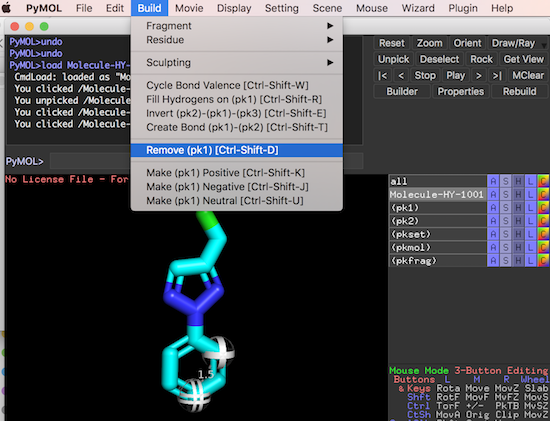
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
"pk1", "pk2", "pk3" and "pk4" Selections
"edit id n1, id n2, id n3, id n4" Commands
►"remove pk*" and "remove_picked" Commands
"unbond pk1, pk2" and "bond pk1, pk2" Commands
"replace new_atom, ..." Replace pk1 with New Atom
"attach new_atom, ..." Attach to pk1 with New Atom
Build Alcohol Molecule with PyMol
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)