Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo - Molecule PNG Image
This section provides a quick introduction on the rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo class, which creates 2D molecule drawings in PNG format.
What Is rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo? - rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo class, together with its base class rdkit.Chem.Draw.rdMolDraw2D.MolDraw2D, provides functionalities to draw molecules in PNG raster image format.
Before playing with the rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo class, let's take a look at how to display PNG images on the screen.
The best way to display PNG image with Python is to use Jupyter Notebook, which uses the IPython kernel and related image display libraries as shown below.
IPython.display.Image A class to create a PNG/JPEG/GIF image object with given image data from a binary string, file, or URL.
- i = IPython.display.Image(d) - Creates a PNG/JPEG/GIF image object with given image data from a binary string.
- i = IPython.display.Image(data=d) - Same as IPython.display.Image(d).
- i = IPython.display.Image(filename=f) - Creates a PNG/JPEG/GIF image object with given image data from an external file.
- i = IPython.display.Image(url=u) - Creates a PNG/JPEG/GIF image object with given image data from a URL.
IPython.display.display(o) - A powerful method that allows you to display IPython image objects and almost any other objects in Notebook. You can use it instead of the Python system print() method to display any object.
The display(o) method is also very special that:
- It is automatically imported in Jupyter and IPython environments. So you can use it anywhere.
- It is the implicit object method in Jupyter and IPython environments. So you can just type o, instead of display(o).
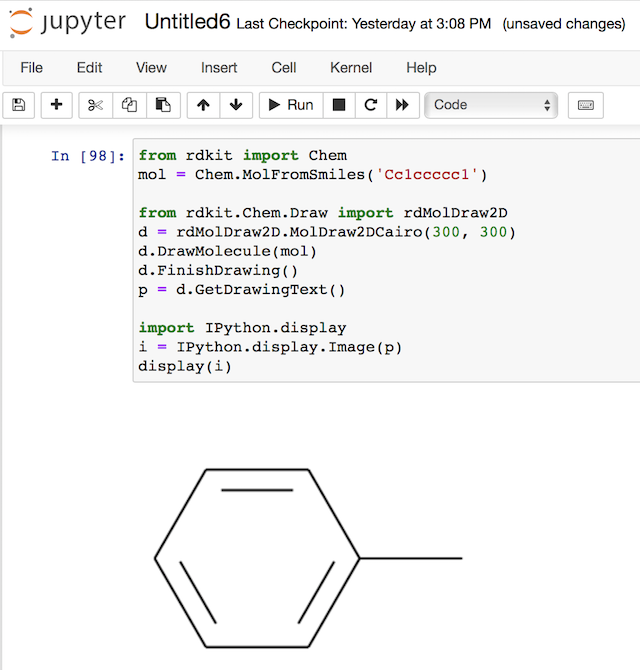
Now, if we combine the power of RDKit and IPython, we can create and display a molecule PNG image very easily in Jupyter Notebook:
from rdkit import Chem
mol = Chem.MolFromSmiles('Cc1ccccc1')
from rdkit.Chem.Draw import rdMolDraw2D
d = rdMolDraw2D.MolDraw2DCairo(300, 300)
d.DrawMolecule(mol)
d.FinishDrawing()
p = d.GetDrawingText()
import IPython.display
i = IPython.display.Image(p)
display(i)
If you want to save the image to a PNG file, you can call the WriteDrawingText(f) method, as shown below:
from rdkit import Chem
mol = Chem.MolFromSmiles('Cc1ccccc1')
from rdkit.Chem.Draw import rdMolDraw2D
d = rdMolDraw2D.MolDraw2DCairo(300, 300)
d.DrawMolecule(mol)
d.FinishDrawing()
d.WriteDrawingText("molecule.png")
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
►rdkit.Chem.Draw - Handle Molecule Images
What Is rdkit.Chem.Draw Module
MolToImage/MolToFile - Molecule PNG Image
rdkit.Chem.Draw.MolDrawing.DrawingOptions Class
rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo - 2D Molecule Drawing
►rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DCairo - Molecule PNG Image
rdkit.Chem.Draw.rdMolDraw2D.MolDraw2DSVG - Molecule SVG Image
rdkit.Chem.Draw.rdMolDraw2D.MolDrawOptions - Drawing Options
Drawing Diagrams with MolDraw2DCairo and MolDraw2DSVG
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction