Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
"obrotamer" - Generate Random Rotational Isomers
This section provides a quick introduction on the 'obrotamer' command provided in Open Babel package to generate random rotational isomers.
What Is "obrotamer" command? - "obrotamer" command is a command line tool provided in the Open Babel package that allows you to Generate Random Rotational Isomers for a given molecule.
Here is the user manual of the "obrotamer" command.
NAME
obrotamer -- generate conformer/rotamer coordinates
SYNOPSIS
obrotamer filename
DESCRIPTION
The obrotamer tool can be used as part of a conformational search by generating
random isomers based on rotating dihedral angles. These rotamers are not
conformers -- that is, obrotamer does not perform geometry optimization
after generating the rotamer structure. The obminimize tool can do geometry
optimization using molecular mechanics.
EXAMPLES
obrotamer baseconformer.sdf >rotamer1.sdf
Generate a random rotational isomer of baseconformer.sdf and write it to rotamer1.sdf
Here is an example of generating random Rotational Isomers with the "obminimize" command:
1. Create a sample molecule with some rotatable bonds:
herong$ obabel "-:CC(N)CCC(C)O" -O sample.sdf --gen2D 1 molecule converted
2. Generate 3 random rotational isomers from the sample molecule:
herong$ obrotamer sample.sdf > sample-isomer-1.sdf Number of rotatable bonds: 3 herong$ obrotamer sample.sdf > sample-isomer-2.sdf Number of rotatable bonds: 3 herong$ obrotamer sample.sdf > sample-isomer-3.sdf Number of rotatable bonds: 3
3. Put the sample molecule and it isomers together in a single SVG picture:
herong$ obabel sample.sdf sample-*.sdf sample-isomers.svg -xcols2 4 molecules converted
4. Compare rotational isomers against the original molecule structure, which is the first one in the first row:
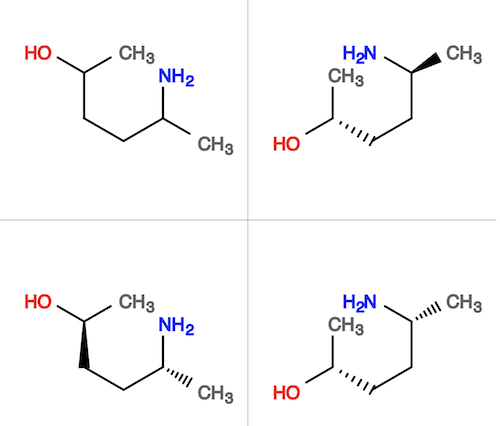
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
►Command Line Tools Provided by Open Babel
List of Open Babel Command Line Tools
"obchiral" - Print Chirality Information
"obconformer" - Generate Best Conformer
"obenergy" - Calculate Molecule Energy
"obfit" - Superimpose Two Molecules
"obgen" - Generate Molecule 3D Structures
"obgrep" - Search Molecules using SMARTS
"obminimize" - Optimize Geometry/Energy of Molecule
"obprobe" - Create Electrostatic Probe Grid
►"obrotamer" - Generate Random Rotational Isomers
"obrotate" - Rotate Dihedral Angles with SMARTS
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction