Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
"obgen" - Generate Molecule 3D Structures
This section provides a quick introduction on the 'obgen' command provided in Open Babel package to generate molecule 3D structure.
What Is "obgen" command? - "obgen" command is a command line tool provided in the Open Babel package that allows you to Generate Molecule 3D Structures. "obgen" command does the same job as the "obabel ... --gen3D" with more options.
Here is the user manual of the "obgen" command.
NAME
obgen -- generate 3D coordinates for a molecule
SYNOPSIS
obgen [OPTIONS] filename
DESCRIPTION
The obgen tool will generate 3D coordinates for molecules in a file (e.g.
multi-molecule SMILES files). The resulting structure will be optimized
using the given forcefield and checked for the lowest-energy conformer
using a Monte Carlo search. Output will be sent to standard output in
the SDF file format.
OPTIONS
If no filename is given, obgen will give all options including the available
forcefields.
-ff forcefield
Select the forcefield
EXAMPLES
View the possible options, including available forcefields:
obgen
Generate 3D coordinates for the molecule(s) in file test.smi:
obgen test.smi
Generate 3D coordinates for the molecule(s) in file test.smi using the UFF forcefield:
obgen -ff UFF test.smi
Here is an example of generating, or converting, a given molecule structure to the best conforming 3D structure using the default force field type:
1. Create a 2D structure of the tyrosine molecule:
herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -O tyrosine.sdf --gen2D 1 molecule converted herong$ obabel tyrosine.sdf tyrosine.svg 1 molecule converted 29 audit log messages
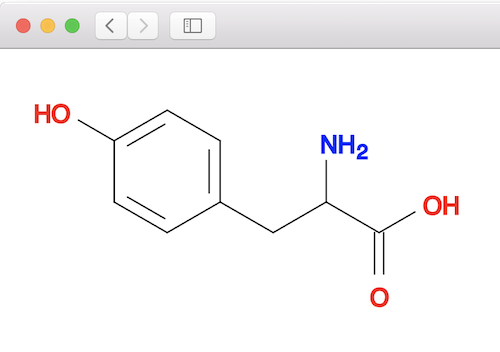
2. Generate, or convert, the 2D structure to 3D structure using the "obgen" command:
herong$ obgen tyrosine.sdf > tyrosine-3d-obgen.sdf
A T O M T Y P E S
IDX TYPE
1 37
2 37
3 37
...
F O R M A L C H A R G E S
IDX CHARGE
1 0.000000
2 0.000000
3 0.000000
...
P A R T I A L C H A R G E S
IDX CHARGE
1 -0.150000
2 -0.150000
3 0.082500
...
S E T T I N G U P C A L C U L A T I O N S
SETTING UP BOND CALCULATIONS...
SETTING UP ANGLE & STRETCH-BEND CALCULATIONS...
SETTING UP TORSION CALCULATIONS...
SETTING UP OOP CALCULATIONS...
SETTING UP VAN DER WAALS CALCULATIONS...
SETTING UP ELECTROSTATIC CALCULATIONS...
S T E E P E S T D E S C E N T
STEPS = 500
STEP n E(n) E(n-1)
------------------------------------
0 124.930 ----
10 43.09167 44.83204
20 29.55319 29.87195
30 20.14396 22.92951
...
STEEPEST DESCENT HAS CONVERGED
W E I G H T E D R O T O R S E A R C H
NUMBER OF ROTATABLE BONDS: 3
NUMBER OF POSSIBLE ROTAMERS: 288
INITIAL WEIGHTING OF ROTAMERS...
GENERATED 250 CONFORMERS
CONFORMER ENERGY
--------------------
1 11.188
2 4.809
3 3.506
.. ...
248 6.265
249 11.152
250 9.037
LOWEST ENERGY: 0.926
S T E E P E S T D E S C E N T
STEPS = 500
STEP n E(n) E(n-1)
------------------------------------
0 0.926 ----
10 0.92061 0.92084
20 0.91410 0.91561
30 0.90976 0.91002
...
STEEPEST DESCENT HAS CONVERGED
herong$ obabel tyrosine-3d-obgen.sdf tyrosine-3d-obgen.svg -d
1 molecule converted
34 audit log messages
You need a 3D structure viewer to see the 3D structure. But you can look at the 2D presentation of the 3D structure.
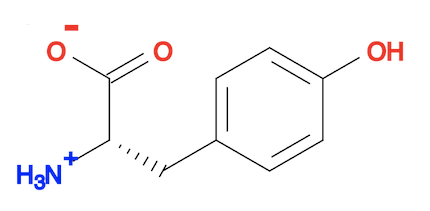
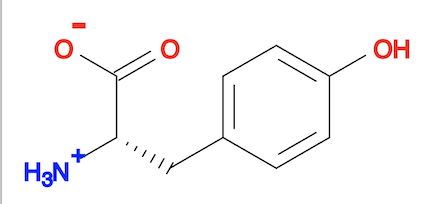
3. You can actually use "obabel ... --gen3D" command to do same job as the "obgen" command. The result is the same as the "obgen" command.
herong$ obabel tyrosine.sdf tyrosine-3d-babel.sdf --gen3D 1 molecule converted herong$ obabel tyrosine-3d-babel.sdf tyrosine-3d-babel.svg -d 1 molecule converted
4. Or you can generate the 3D structure from the SMILES string directly using the "obabel" command. The result is almost the same as the "obgen" command.
herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -O tyrosine-3d-def.sdf --gen3D 1 molecule converted herong$ obabel tyrosine-3d-def.sdf tyrosine-3d-def.svg -d 1 molecule converted 34 audit log messages
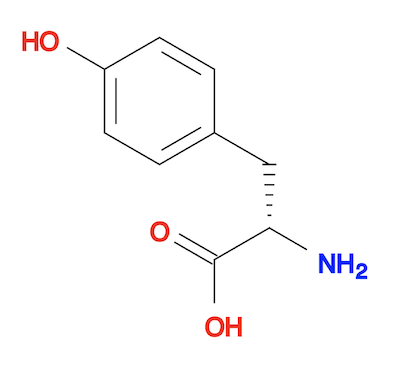
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
►Command Line Tools Provided by Open Babel
List of Open Babel Command Line Tools
"obchiral" - Print Chirality Information
"obconformer" - Generate Best Conformer
"obenergy" - Calculate Molecule Energy
"obfit" - Superimpose Two Molecules
►"obgen" - Generate Molecule 3D Structures
"obgrep" - Search Molecules using SMARTS
"obminimize" - Optimize Geometry/Energy of Molecule
"obprobe" - Create Electrostatic Probe Grid
"obrotamer" - Generate Random Rotational Isomers
"obrotate" - Rotate Dihedral Angles with SMARTS
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction