Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
"obfit" - Superimpose Two Molecules
This section provides a quick introduction on the 'obfit' command provided in Open Babel package to superimpose two molecules.
What Is "obfit" Command? - "obfit" command is a command line tool provided in the Open Babel package that allows you to Superimpose Two Molecules.
Here is the user manual of the "obfit" command.
NAME
obfit -- superimpose two molecules based on a pattern
SYNOPSIS
obfit SMARTS-pattern fixed-file outfile
DESCRIPTION
Superimpose two molecules using a quaternion fit. The atoms used to fit
the two molecules are defined by the SMARTS pattern given by the user. It
is useful to align congeneric series of molecules on a common structural
scaffold for 3D-QSAR studies. It can also be useful for displaying the
results of conformational generation.
Any molecules matching the supplied SMARTS pattern will be rotated and
translated to provide the smallest possible RMSD between the matching regions.
If a molecule does not match the SMARTS pattern, it will be output with no
transformation.
EXAMPLES
Align all the molecules in 'testmv.sdf' on a single molecule of 'testref.sdf'
by superimposing them on its N-methylpiperidyl portion (and outputting a
new SD file to the standard output):
obfit '[nh]1c2c(=O)n(C)c(=O)n(C)c2cc1' testref.sdf testmv.sdf
Here is an example of superimposing, or aligning, molecule structures against a reference molecule structures using the "obift" command:
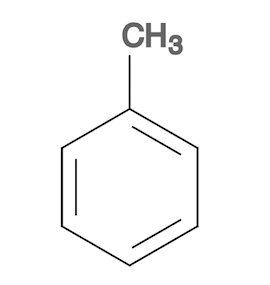
1. Create a toluene molecule structure as the reference molecule with a molecule editor so that you can keep the extra carbon on top of the benzene ring. Save the the toluene molecule structure in toluene.sdf.
2. Convert toluene.sdf to toluene.svg to see the toluene molecule structure.
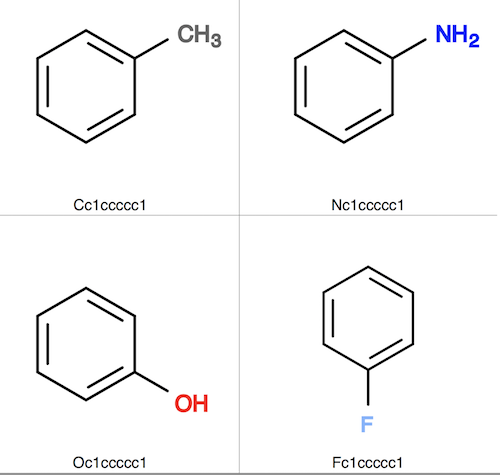
3. Create 4 more molecule structures similar to the toluene molecule structure, by rotating the ring and replacing C with N, O, and F. Save them in a single file, fit-input.sdf.
4. Convert fit-input.sdf to fit-input.svg to see those similar molecule structures.
5. Now fit those those similar molecule structures against the toluene molecule structure with the help of SMARTS pattern "c1cc(*)ccc1".
herong$ obfit 'c1cc(*)ccc1' toluene.sdf fit-input.sdf > fit-output.sdf RMSD: 0.000032 RMSD: 0.000032 RMSD: 0.000032 RMSD: 0.000000
6. Convert fit-output.sdf to fit-output.svg to see that all similar molecule structures are superimposed (aligned) to the toluene molecule structure.
herong$ obabel fit-output.sdf fit-output.svg 4 molecules converted 110 audit log messages
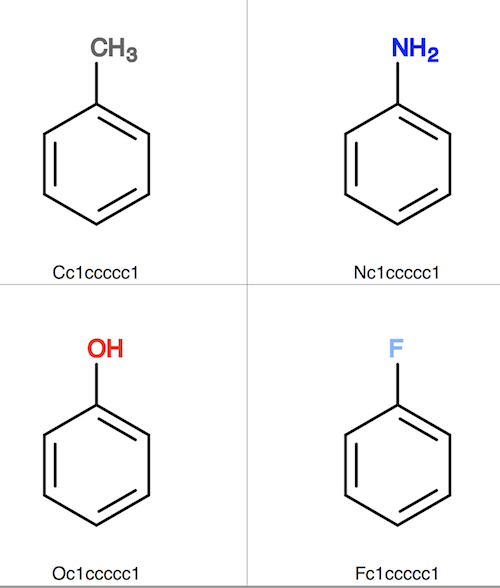
As you can see, "obfit" did a good job to superimpose or align structures of all similar molecules to the given reference molecule structure.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
►Command Line Tools Provided by Open Babel
List of Open Babel Command Line Tools
"obchiral" - Print Chirality Information
"obconformer" - Generate Best Conformer
"obenergy" - Calculate Molecule Energy
►"obfit" - Superimpose Two Molecules
"obgen" - Generate Molecule 3D Structures
"obgrep" - Search Molecules using SMARTS
"obminimize" - Optimize Geometry/Energy of Molecule
"obprobe" - Create Electrostatic Probe Grid
"obrotamer" - Generate Random Rotational Isomers
"obrotate" - Rotate Dihedral Angles with SMARTS
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction