Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
"obconformer" - Generate Best Conformer
This section provides a quick introduction on the 'obconformer' command provided in Open Babel package to generate best conformer.
What Is "obconformer" Command? - "obconformer" command is a command line tool provided in the Open Babel package that allows you to generate the best conformer from a base molecule.
Here is the user manual of the "obconformer" command.
NAME
obconformer -- generate conformer coordinates
SYNOPSIS
obconformer # of conformers # of optimization steps filename
DESCRIPTION
The obconformer tool can be used as part of a conformational study by
generating random conformers using a Monte Carlo search. The best conformer
out of the batch of conformers will be output, after taking the supplied number
of geometry optimization steps. By default, obconformer uses the MMFF94
force field.
EXAMPLES
obconformer 250 100 baseconformer.sdf >rotamer1.sdf
Generate the best conformer (out of 250) of baseconformer.sdf and write it
to rotamer1.sdf after 100 geometry
optimization steps
Here is an example of generating best conformer using the "obconformer" command.
herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o sdf -O tyrosine.sdf --gen2D
1 molecule converted
herong$ obconformer 250 100 tyrosine.sdf > conformer.sdf
A T O M T Y P E S
IDX TYPE
1 37
2 37
3 37
4 37
5 37
6 37
7 4
8 2
9 41
10 32
11 32
12 8
13 35
F O R M A L C H A R G E S
IDX CHARGE
1 0.000000
2 0.000000
3 0.000000
4 0.000000
5 0.000000
6 0.000000
7 0.000000
8 0.000000
9 0.000000
10 -0.500000
11 -0.500000
12 0.000000
13 -1.000000
P A R T I A L C H A R G E S
IDX CHARGE
1 0.000000
2 0.000000
3 -0.171000
4 0.000000
5 0.000000
6 0.073000
7 -0.138000
8 -0.067000
9 1.050000
10 -0.900000
11 -0.900000
12 -0.118000
13 -0.829000
S E T T I N G U P C A L C U L A T I O N S
SETTING UP BOND CALCULATIONS...
USING EMPIRICAL RULE FOR BOND STRETCHING 8-12 (IDX)...
SETTING UP ANGLE & STRETCH-BEND CALCULATIONS...
USING EMPIRICAL RULE FOR ANGLE BENDING 6-7-8 (IDX)...
USING EMPIRICAL RULE FOR ANGLE BENDING FORCE CONSTANT 6-7-8 (IDX)...
USING EMPIRICAL RULE FOR ANGLE BENDING FORCE CONSTANT 7-8-9 (IDX)...
USING EMPIRICAL RULE FOR ANGLE BENDING FORCE CONSTANT 7-8-12 (IDX)...
USING EMPIRICAL RULE FOR ANGLE BENDING FORCE CONSTANT 9-8-12 (IDX)...
SETTING UP TORSION CALCULATIONS...
SETTING UP OOP CALCULATIONS...
SETTING UP VAN DER WAALS CALCULATIONS...
SETTING UP ELECTROSTATIC CALCULATIONS...
W E I G H T E D R O T O R S E A R C H
NUMBER OF ROTATABLE BONDS: 3
NUMBER OF POSSIBLE ROTAMERS: 480
INITIAL WEIGHTING OF ROTAMERS...
GENERATED 250 CONFORMERS
CONFORMER ENERGY
--------------------
1 90.092
2 90.185
3 90.501
4 92.225
5 90.318
...
248 90.255
249 90.081
250 90.106
LOWEST ENERGY: 90.077
C O N J U G A T E G R A D I E N T S
STEPS = 100
STEP n E(n) E(n-1)
--------------------------------
1 90.077 90.077
10 90.077 90.076
20 90.077 90.077
30 90.077 90.076
40 90.076 90.076
50 90.077 90.076
60 90.076 90.076
70 90.076 90.076
80 90.076 90.075
90 90.077 90.076
100 90.076 90.075
Now generate SVG pictures for both the conformer and the base molecule.
herong$ obabel tyrosine.sdf tyrosine.svg 1 molecule converted 29 audit log messages herong$ obabel conformer.sdf conformer.svg 1 molecule converted 33 audit log messages
Open tyrosine.svg in a Web browser to see the base molecule structure:
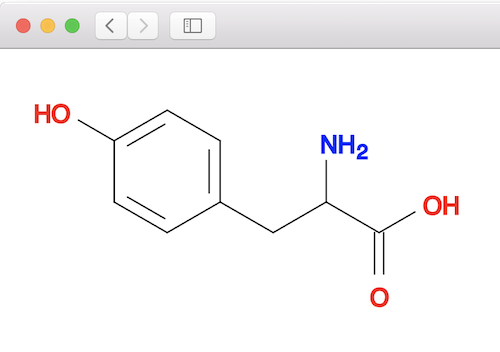
Open conformer.svg in a Web browser to compare the best conformer with the base molecule:
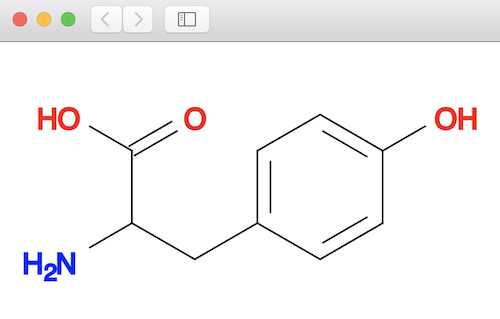
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
►Command Line Tools Provided by Open Babel
List of Open Babel Command Line Tools
"obchiral" - Print Chirality Information
►"obconformer" - Generate Best Conformer
"obenergy" - Calculate Molecule Energy
"obfit" - Superimpose Two Molecules
"obgen" - Generate Molecule 3D Structures
"obgrep" - Search Molecules using SMARTS
"obminimize" - Optimize Geometry/Energy of Molecule
"obprobe" - Create Electrostatic Probe Grid
"obrotamer" - Generate Random Rotational Isomers
"obrotate" - Rotate Dihedral Angles with SMARTS
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction