Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
rdkit.Chem.rdFMCS - Maximum Common Substructure
This section provides a quick introduction on rdkit.Chem.rdFMCS module in RDKit library that provides functionalities to find the MCS (Maximum Common Substructure) of a set of molecules.
What Is rdkit.Chem.rdFMCS Module? rdkit.Chem.rdFMCS module in RDKit library provides functionalities to find the MCS (Maximum Common Substructure) of a set of molecules.
rdkit.Chem.rdFMCS module contains one main method and one main class:
r = rdkit.Chem.rdFMCS.FindMCS(ms) - Finds the MCS for a set of molecules and returns it as a rdkit.Chem.rdFMCS.MCSResult object. This method uses the following algorithm:
best_substructure = None
pick one structure in the set as query, all other as targets
for each substructure in the query:
convert into a SMARTS string based on the desired match properties
if SMARTS pattern exists in all of the targets:
then it is a common substructure
keep track of the maximum of such substructure
rdkit.Chem.rdFMCS.MCSResult - Represents the MCS returned from the FindMCS() method. It contains the following properties:
- r.canceled - If True, the MCS calculation did not finish
- r.numAtoms - Number of atoms in MCS
- r.numBonds - Number of bonds in MCS
- r.queryMol - Query molecule for the MCS
- r.smartsString - SMARTS string for the MCS
Here is a nice short Jupyter Notebook example of finding the MCS using RDKit library.
import rdkit.Chem ms = ["CCC1CC2C1CN2", "C1CC2C1CC2"] ms = list(map(rdkit.Chem.MolFromSmiles, ms)) i = rdkit.Chem.Draw.MolsToGridImage(ms, subImgSize=(150,150)) r = rdkit.Chem.rdFMCS.FindMCS(ms) display(i, r.smartsString) rdkit.Chem.Draw.MolToImage(r.queryMol, size=(120,120))
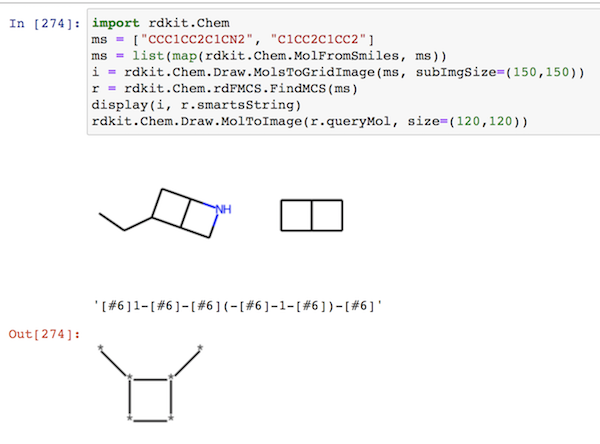
For more information on MCS with RDKit, see "T006 · Maximum common substructure" at https://projects.volkamerlab.org/teachopencadd/index.html.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
►Molecule Substructure Search with RDKit
RDKit m.HasSubstructMatch(s) - Substructure Match
RDKit GenerateDepictionMatching2DStructure(m, s) - Substructure Orientation
RDKit rdMolDraw2D.PrepareAndDrawMolecule - Substructure Highlight
RDKit Substructure Search with SMARTS
►rdkit.Chem.rdFMCS - Maximum Common Substructure
rdkit.Chem.rdSubstructLibrary - Substructure Library
Substructure Library in Binary and SMILES Formats
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction