Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
Hash over Double Bond by Open Babel
This section provides a tutorial example demonstrating how Open Babel 'correcting' wedge-hash bond representations of stereo centers from molecule input data.
If a stereo center has a double bond, and Open Babel decides to bend it for representing the chirality, the double bond will be displayed as a wedge or hash bond.
Here is an example, where Open Babel replaces the double bond sign with the hash sign to represent the chirality for the first molecule structure:
herong$ obabel '-:OC([P@](=O)(NC)O)' -O chirality-1.smi herong$ obabel '-:OC([P@](=OCC)(NC)O)' -O chirality-2.smi herong$ obabel chirality-1.smi chirality-2.smi -O output.svg herong$ chrome output.svg
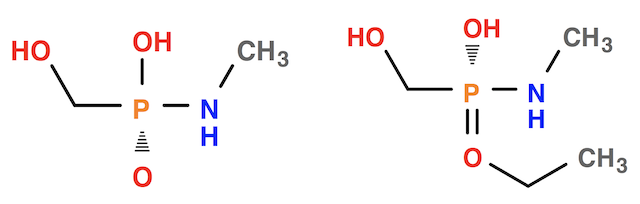
If you look at the first molecule structure, you can see that the P=O branch is the most simple branch of the stereo center. So Open Babel decided to bend P=O with a hash sign.
If you look at the second molecule structure, you can see that the PO branch is the most simple branch of the stereo center. Open Babel decided to bend PO with a hash sign.
In other words, Open Babel selects the most simple branch and bends it with a wedge or hash sign to represent the chirality. It doesn't care if selected branch is a single bond or double bond.
By the way, phosphorus (P) is a multivalent atom and can form +3 and +5 cations. So it can have 3 or 5 pairs of bonds in stable state.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
►Stereochemistry with Open Babel
Read Stereoinformation from Input with Open Babel
Stereo Perception Performed by Open Babel
Write Stereoinformation to Output by Open Babel
Wedge-Hash Bond Changed by Open Babel
Hash Bond with Solid Line by Open Babel
►Hash over Double Bond by Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction