Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
Use StoneMIND Collector Web Interface
This section provides a tutorial example on how to use the StoneMIND Collector Web Interface to scan an entire patent PDF document of 181 pages and recognize all molecules using 'IUPAC' and 'OCSR' methods.
StoneMIND Collector Interface offers additional functionalities to do batch extraction of patents or essays. Here is what I did to try
1. Go to StoneMIND Collector Website at https://www.stonewise.cn/mol_product.
2. Click the "Web Interface" button. I see the signin/signup screen in Chinese.
3. Click "Signup" and fill in the form and click "Submit". I see the StoneMIND Collector Web interface.
4. Click Knowledge Base > Data Extraction. I see an empty project list.
5. Click "Create Project" and enter "Test" as the project name. I see no tasks in the new project.
6. Click "Create Task". I see a task window with 2 options: "Upload PDF" and "Paten Number".
7. Select "Patent Number" and enter "WO2001000214A1". I see a new task created. StoneMIND Collector is smart to find the PDF file of the given patent number from the Internet automatically.
8. Click "Extract" on the task and select both "IUPAC" and "OCSR" methods. I see that StoneMIND Collector starts to scan the PDF file and extracts molecule structures.
11. Click "View" after the extraction process is completed, which may take some time. I see 188 molecules extracted from OCSR method and 189 from IUPAC method.
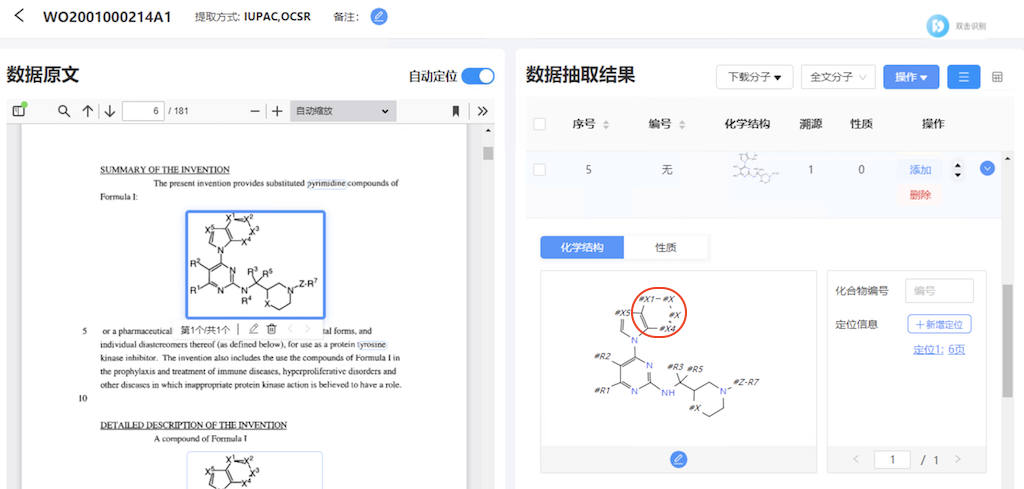
12. Go to page 6 in the PDF panel on the left. And click the first molecule diagram. I see the resulting molecule structure displayed on the right. It looks more accurate than the result done by StoneMIND Collector client. The top ring is complete and only mislabeled X2 and X3 atoms.
13. Inspect each extracted molecule and correct any mistakes as you see.
14. Click "Download" to save all extracted molecules to a local file.
Here is a summary of my StoneMIND Collector Web interface task:
Patent Number: WO2001000214A1 Patent Year: 2001 PDF Pages: 181 Molecules by OCSR: 188 Molecules by IUPAC: 189 Extraction Time in Minutes: 74 Seconds per Page: 25
Conclusions:
- StoneMIND Collector Web is easy to use.
- It is slow, about 25 seconds per page.
- It is pretty accurate.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
►OCSR (Optical Chemical Structure Recognition)
StoneMIND Collector - Information Extraction System
Install StoneMIND Collector Client on Windows
Use StoneMIND Collector on Windows
Stop StoneMIND Collector on Windows
►Use StoneMIND Collector Web Interface
AlphaFold - Protein Structure Prediction