Cheminformatics Tutorials - Herong's Tutorial Examples - v2.03, by Herong Yang
Morgan Fingerprint Generator in RDKit for FCFP
This section provides a tutorial on how to generate FCFP fingerprints with the Morgan Fingerprint Generator in the RDKit library.
The Morgan fingerprint generator in RDKit also supports FCFP (Functional-Class Fingerprints) by using the "useFeatures=True" when calling rdkit.Chem.rdMolDescriptors.GetMorganFingerprint(), rdkit.Chem.rdMolDescriptors.GetMorganFingerprintAsBitVect(), or rdkit.Chem.rdMolDescriptors.GetHashedMorganFingerprint().
When generating FCFP fingerprints, RDKit replaces initial identifiers generate from pharmacophoric properties (functional-class invariants) instead of atom invariants, as described in the "FCFP (Functional-Class Fingerprints) Method" section in this book.
RDKit generates initial identifiers as 6-bit integers with each bit to represent one of the following pharmacophoric conditions on each atom node.
- Bit 0: Hydrogen bond donor.
- Bit 1: Hydrogen bond acceptor.
- Bit 2: Aromatic.
- Bit 3: Halogen.
- Bit 4: Basic group.
- Bit 5: Acidic group.
By the way, those initial identifiers are also referred as feature identifiers, since they represent pharmacophoric features of the molecule.
Now let's verify our understanding with some tests.
1. The first test is the FCFP fingerprint of a Benzene ring using radius=0 to keep only initial identifiers in the fingerprint.
from rdkit.Chem import AllChem
from rdkit.DataStructs.cDataStructs import UIntSparseIntVect
bitInfo = {}
mol = AllChem.MolFromSmiles('c1ccccc1')
fp = AllChem.GetMorganFingerprint(mol, 0, useFeatures=True, bitInfo=bitInfo)
display(UIntSparseIntVect.GetNonzeroElements(fp))
print(bitInfo)
# output:
{4: 6}
{4: ((0, 0), (1, 0), (2, 0), (3, 0), (4, 0), (5, 0))}
The output shows all 6 atom nodes have the same initial identifier 4, or 0b000100. Only bit 2 is turned on as all nodes are on an aromatic ring.
2. The second test uses a simple carboxylic acid, CC(=O)O.
from rdkit.Chem import AllChem
from rdkit.DataStructs.cDataStructs import UIntSparseIntVect
bitInfo = {}
mol = AllChem.MolFromSmiles('CC(=O)O')
fp = AllChem.GetMorganFingerprint(mol, 0, useFeatures=True, bitInfo=bitInfo)
display(UIntSparseIntVect.GetNonzeroElements(fp))
print(bitInfo)
# output:
{0: 1, 1: 1, 2: 1, 32: 1}
{0: ((0, 0),), 1: ((3, 0),), 2: ((2, 0),), 32: ((1, 0),)}
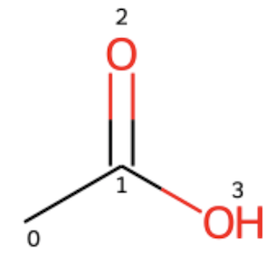
The output shows 4 atom nodes have different initial identifiers:
- Atom 0: 0 = 0b000000, with no pharmacophoric property.
- Atom 1: 32 = 0b100000, with bit 5 turned on representing an acidic group.
- Atom 2: 2 = 0b000010, with bit 1 turned on representing an Hydrogen bond acceptor.
- Atom 3: 1 = 0b000001, with bit 0 turned on representing an Hydrogen bond donor.
You can use the code below to view the structure of CC(=O)O with atom indexes displayed.
from rdkit.Chem.Draw import IPythonConsole
IPythonConsole.drawOptions.addAtomIndices = True
IPythonConsole.drawOptions.addBondIndices = False
mol = Chem.MolFromSmiles('CC(=O)O')
display(mol)
2. Now let's try the radius=1 for carboxylic acid, CC(=O)O.
from rdkit.Chem import AllChem
from rdkit.DataStructs.cDataStructs import UIntSparseIntVect
bitInfo = {}
mol = AllChem.MolFromSmiles('CC(=O)O')
fp = AllChem.GetMorganFingerprint(mol, 1, useFeatures=True, bitInfo=bitInfo)
display(UIntSparseIntVect.GetNonzeroElements(fp))
print(bitInfo)
# output:
{ 0: 1,
1: 1,
2: 1,
32: 1,
605976925: 1,
3205495832: 1,
3205495901: 1,
3205496734: 1}
{ 0: ((0, 0),),
1: ((3, 0),),
2: ((2, 0),),
32: ((1, 0),),
605976925: ((1, 1),),
3205495832: ((2, 1),),
3205495901: ((0, 1),),
3205496734: ((3, 1),)}
The output shows 2 sets of identifiers, one for the initial round and one for the second round in the Morgan algorithm.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
►Morgan Fingerprint Generator in RDKit
What Is Morgan Fingerprint Generator in RDKit
GetMorganFingerprint() Method in RDKit
Impact of 'radius' on GetMorganFingerprint()
Impact of 'useCounts' on GetMorganFingerprint()
Impact of 'invariants' on GetMorganFingerprint()
Impact of 'useBondTypes' on GetMorganFingerprint()
Impact of 'fromAtoms' on GetMorganFingerprint()
GetMorganFingerprintAsBitVect() Method in RDKit
Impact of 'nBits' on GetMorganFingerprintAsBitVect()
GetHashedMorganFingerprint() Method in RDKit
Impact of 'nBits' on GetHashedMorganFingerprint()
GetMorganGenerator() Method in RDKit
►Morgan Fingerprint Generator in RDKit for FCFP
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction