Cheminformatics Tutorials - Herong's Tutorial Examples - v2.01, by Herong Yang
Impact of 'branchedPaths' on RDKFingerprint()
This section provides a tutorial example on impact of the 'branchedPaths' option on fingerprint generation with rdkit.Chem.rdmolops.RDKFingerprint() function.
The 'branchedPaths' option in the rdkit.Chem.rdmolops.RDKFingerprint() function call allows you to control whether subgraphs with branched paths should be used to turn on bits in the fingerprint. If branchedPaths=True is used, subgraphs with branched paths are included in the fingerprint generation process. If branchedPaths=False is used, subgraphs with branched paths are eliminated in the fingerprint generation process.
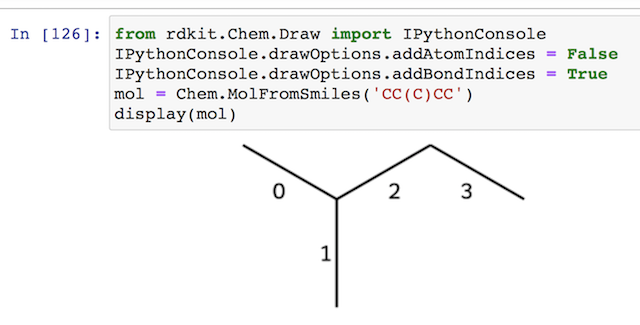
1. For example, "CC(C)CC" is a molecule with 2 branches as displayed in this 2D structure on Jupyter Notebook with the following code. Bond indices are displayed to help us identifying subgraphs.
from rdkit.Chem.Draw import IPythonConsole
IPythonConsole.drawOptions.addAtomIndices = False
IPythonConsole.drawOptions.addBondIndices = True
mol = Chem.MolFromSmiles('CC(C)CC')
display(mol)
2. If we use the "branchedPaths=True" option, the fingerprint will be generated from 5 unique subgraphs.
atomBits = []
bitInfo = {}
mol = Chem.MolFromSmiles('CC(C)CC')
fp = Chem.RDKFingerprint(mol,
fpSize=64, nBitsPerHash=1, branchedPaths=True,
atomBits=atomBits, bitInfo=bitInfo)
print(fp.ToBitString())
print(fp.GetNumOnBits())
print(atomBits)
print(bitInfo)
# output:
0000000000000000000001000000100000010000000100000000000000000100
5
[[28, 21, 43, 61, 35], [28, 21, 43, 61, 35], [28, 21, 61, 43, 35], ...]
{ 21: [[0, 2], [0, 1], [1, 2], [2, 3]],
28: [[0], [1], [2], [3]],
35: [[0, 2, 3, 1]],
43: [[0, 2, 3], [1, 2, 3]],
61: [[0, 2, 1]]
}
As you can see from the output, there are 2 branched subgraphs included in the fingerprint: bit 35 with bonds of [0, 2, 3, 1], and bit 61 with bonds of [0, 2, 1].
3. Now if we use the "branchedPaths=False" option, the fingerprint will be generated from only 3 non-branched unique subgraphs.
atomBits = []
bitInfo = {}
mol = Chem.MolFromSmiles('CC(C)CC')
fp = Chem.RDKFingerprint(mol,
fpSize=64, nBitsPerHash=1, branchedPaths=False,
atomBits=atomBits, bitInfo=bitInfo)
print(fp.ToBitString())
print(fp.GetNumOnBits())
print(atomBits)
print(bitInfo)
# output:
0000000000000000000001000000100000000000000100000000000000000000
3
[[28, 21, 43], [28, 21, 43], [28, 21, 43], [28, 21, 43], [28, 21, 43]]
{ 21: [[0, 1], [0, 2], [2, 3], [1, 2]],
28: [[0], [1], [2], [3]],
43: [[0, 2, 3], [1, 2, 3]]
}
4. Without branched subgraphs, 2 different molecules, "CC(C)CC" and "CCCC" will have identical fingerprints.
mol = Chem.MolFromSmiles('CC(C)CC')
fp1 = Chem.RDKFingerprint(mol,
fpSize=64, nBitsPerHash=1, branchedPaths=False,
atomBits=atomBits, bitInfo=bitInfo)
print(fp1.ToBitString())
mol = Chem.MolFromSmiles('CCCC')
fp2 = Chem.RDKFingerprint(mol,
fpSize=64, nBitsPerHash=1, branchedPaths=False,
atomBits=atomBits, bitInfo=bitInfo)
print(fp2.ToBitString())
# output:
0000000000000000000001000000100000000000000100000000000000000000
0000000000000000000001000000100000000000000100000000000000000000
Conclusion: We should keep using "branchedPaths=True" option so that differences branched subgraphs and non-branched will be reflected the fingerprint.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
►Daylight Fingerprint Generator in RDKit
What Is Daylight Fingerprint Generator in RDKit
RDKFingerprint() Method in RDKit
Impact of 'useBondOrder' on RDKFingerprint()
►Impact of 'branchedPaths' on RDKFingerprint()
Impact of 'maxPath' on RDKFingerprint()
Impact of 'fpSize' on RDKFingerprint()
Impact of 'tgtDensity' on RDKFingerprint()
Impact of 'nBitsPerHash' on RDKFingerprint()
UnfoldedRDKFingerprintCountBased() Method in RDKit
GetRDKitFPGenerator() Method in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction