Cheminformatics Tutorials - Herong's Tutorial Examples - v2.01, by Herong Yang
"obabel ... --gen2D" - Calculated 2D Coordinates
This section provides tutorial examples on how to use Open Babel command '--gen2D' option to calculate 2D coordinates of each atom in the given molecule.
"--gen2D" is an "obabel" command option that tells Open Babel to calculate 2D coordinates of each atom in the given molecule. There are 3 possible ways of using the "--gen2D" option.
1. "--gen2D" Implicitly Applied - If the output format like SVG that requires the molecule to be presented a 2D space, and the input data does not contain 2D coordinates, Open Babel will automatically apply the "--gen2D" option and calculate 2D coordinates of each atom in the molecule.
For example, the following "obabel" command converts a benzene molecule from SMILES format to SVG format without "--gen2D" option. 2D coordinates are calculated automatically.
herong$ obabel -:c1ccccc1 -O benzene.svg
2. "--gen2D" Explicitly Suggested - If the output format like SDF that is capable to record 2D coordinates but not required, and the input data does not contain 2D coordinates, Open Babel will not apply the "--gen2D" option and give a warning message.
For example, the following command converts a benzene molecule from SMILES format to SDF format without "--gen2D" option. 2D coordinates are not calculated.
herong$ obabel -:c1ccccc1 -O benzene-without-gen2D.sdf
==============================
*** Open Babel Warning in WriteMolecule
No 2D or 3D coordinates exist. Stereochemical information will be
stored using an Open Babel extension. To generate 2D or 3D coordinates
instead use --gen2D or --gen3D.
1 molecule converted
herong$ more benzene-without-gen2D.sdf
OpenBabel10092109552D
6 6 0 0 0 0 0 0 0 0999 V2000
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 6 2 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
M END
herong$ obabel benzene-without-gen2D.sdf -O benzene.svg
Note that benzene-without-gen2D.sdf is still a valid molecule file. You can structure the benzene structure according to the list of atoms and bonds.
If you want to record 2D coordinates in the SDF output file, you can specify the suggested "--gen2D" option.
herong$ obabel -:c1ccccc1 -O benzene-with-gen2D.sdf --gen2D
1 molecule converted
herong$ more benzene-with-gen2D.sdf
OpenBabel10092110102D
6 6 0 0 0 0 0 0 0 0999 V2000
-0.8660 -0.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7321 -0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7321 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8660 1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0000 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 6 2 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
M END
$$$$
3. "--gen2D" Used to Recalculate - When input data already contains 2D coordinates, you can still specify the "--gen2D" option to recalculate them.
For example, the following command converts a guanosine molecule from SDF format to SVG format with "--gen2D" option. 2D coordinates are recalculated.
herong$ obabel guanosine-poor.sdf -O guanosine-with-gen2D.svg --gen2D 1 molecule converted
As a comparison, the following command converts the same SDF file to SVG format without "--gen2D" option. The original 2D coordinates are used.
herong$ obabel guanosine-poor.sdf -O guanosine-without-gen2D.svg 1 molecule converted
The following picture shows the molecule 2D structure with 2D coordinates recalculated by Open Babel on the left, and the 2D structure of the original 2D coordinates on the right.
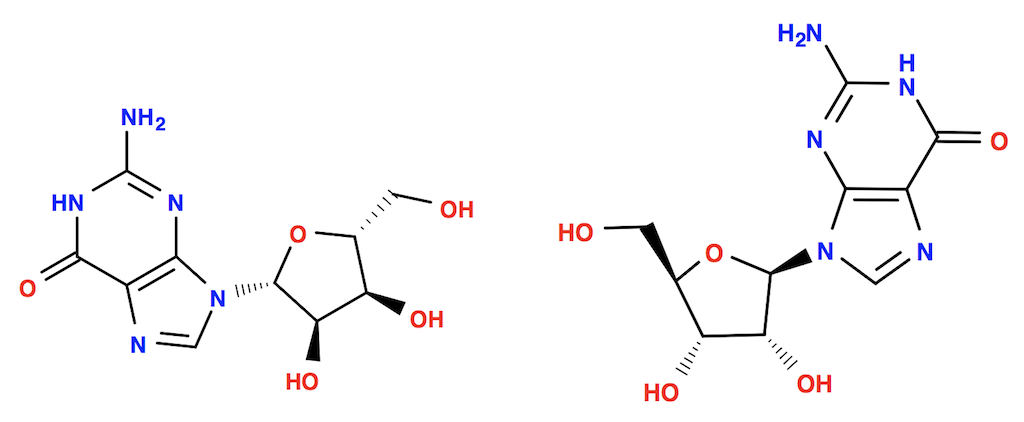
As you can see that Open Babel 2D coordinates recalculation does improve the structure quality. But it also changes the orientation.
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
►Using Open Babel Command: "obabel"
"obabel -i ..." - Input Data Format and Source
"obabel -o ... -O" - Output Data Format and Destination
"obabel -... --..." - Generic Conversion Options
"obabel" Command Option Argument Syntax
►"obabel ... --gen2D" - Calculated 2D Coordinates
"obabel ... -f # -l #" - Split Large Molecule File
"obabel -h/-d" - Add/Remove Hydrogens in Molecule Data
"obabel --append ..." - Calculate Molecule Properties
"obabel -L formats" - List of File Formats Supported
"obabel -a..." - Extra Options for Input Reading
"obabel -x..." - Extra Options for Output Writing
"obabel" vs. "babel" Open Babel Commands
Generating SVG Pictures with Open Babel
Substructure Search with Open Babel
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction