Cheminformatics Tutorials - Herong's Tutorial Examples - v2.01, by Herong Yang
"obabel -s ..." Command - Substructure Search
This section provides a tutorial example on how to use 'obabel ... -s ...' command to do substructure search with Open Babel.
Substructure search is a common task in cheminformatics studies. The objective of substructure search is to evaluate whether or not a given molecule structure contains a given substructure pattern.
You can do substructure search with Open Babel using the "obabel ... -s ..." command with the following syntax:
babel input_section output_section -s smarts_string
The smarts_string specifies a SMARTS string that represents a molecule pattern. "obabel" command will apply this pattern to each molecule in the input data source. If the the pattern matches a substructure of the molecule, it will be written to the output. Otherwise, it will be skipped.
Substructure search is also called substructure filtering or substructure matching.
Here are some examples of substructure matching with a single atom as the molecule pattern:
herong$ # methane molecule contains an aliphatic carbon herong$ obabel -:C -o smiles -s C C 1 molecule converted herong$ # methane molecule contains no aromatic carbon herong$ obabel -:C -o smiles -s c 0 molecules converted herong$ # benzene molecule contains an aromatic carbon herong$ obabel -:c1ccccc1 -o smiles -s c c1ccccc1 1 molecule converted herong$ # benzene molecule contains no aliphatic carbon herong$ obabel -:c1ccccc1 -o smiles -s C 0 molecules converted
Here is another group of examples of substructure matching with a single bond as the molecule pattern:
herong$ # tyrosine molecule contains a two-aromatic-carbon bond herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s cc c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted herong$ # tyrosine molecule contains an aromatic-carbon-aliphatic-carbon bond herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s cC c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted herong$ # tyrosine molecule contains no aromatic-carbon-nitrogen bond herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s cN 0 molecules converted herong$ # tyrosine molecule contains an aliphatic-carbon-nitrogen bond herong$ obabel "-:c1cc(ccc1CC(C(=O)O)N)O" -o smiles -s CN c1cc(ccc1CC(C(=O)O)N)O 1 molecule converted
You can validate the above matching result by looking at the tyrosine molecule structure below:
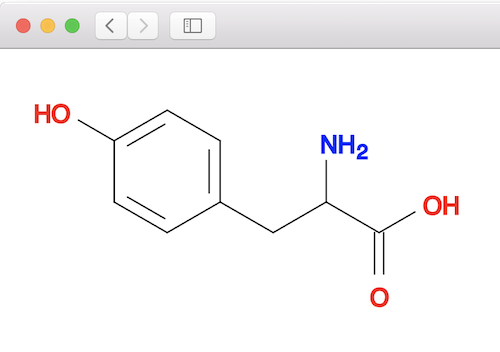
Table of Contents
SMILES (Simplified Molecular-Input Line-Entry System)
Open Babel: The Open Source Chemistry Toolbox
Using Open Babel Command: "obabel"
Generating SVG Pictures with Open Babel
►Substructure Search with Open Babel
►"obabel -s ..." Command - Substructure Search
Substructure Search with Wildcard Atom "*"
Substructure Search with Wildcard Bond "~"
Substructure Search with SMARTS Expressions
Similarity Search with Open Babel
Fingerprint Index for Fastsearch with Open Babel
Stereochemistry with Open Babel
Command Line Tools Provided by Open Babel
RDKit: Open-Source Cheminformatics Software
rdkit.Chem.rdchem - The Core Module
rdkit.Chem.rdmolfiles - Molecular File Module
rdkit.Chem.rdDepictor - Compute 2D Coordinates
rdkit.Chem.Draw - Handle Molecule Images
Molecule Substructure Search with RDKit
rdkit.Chem.rdmolops - Molecule Operations
Daylight Fingerprint Generator in RDKit
Morgan Fingerprint Generator in RDKit
RDKit Performance on Substructure Search
Introduction to Molecular Fingerprints
OCSR (Optical Chemical Structure Recognition)
AlphaFold - Protein Structure Prediction